Scientists Can Use CRISPR to Store Images and Movies in Bacteria
It’s a step toward turning microbes into living hard drives.
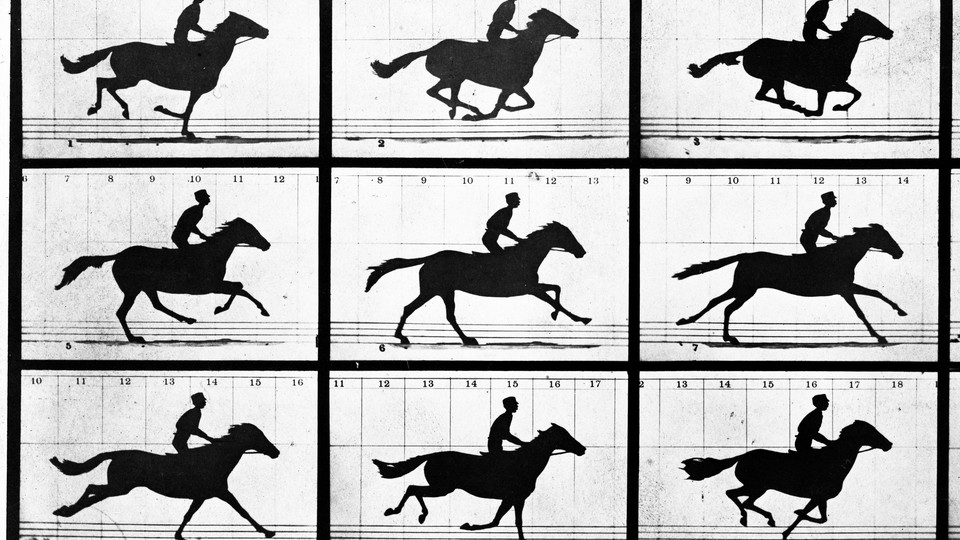
In 1872, Eadweard Muybridge captured a series of photos of a running horse. His images settled a debate about whether the animal ever lifted all four feet off the ground at once—it did. They also formed the basis of one of the first motion pictures. And now, Seth Shipman, from Harvard Medical School, has immortalized the running horse in a new and very different medium: the genomes of bacteria.
Shipman encoded a GIF of Muybridge’s horse into DNA, and then inserted those strands into living microbes using CRISPR. This technique is best known as a tool for editing genes by cutting strands of DNA at precise locations. But it has another trait that’s often overlooked: It’s an amazing tool for recording information. Shipman effectively turned bacteria into living hard drives.
CRISPR was invented billions of years ago, as a way for bacteria to defend themselves against viruses. The bacteria grab the DNA of invading viruses, incorporating it into their own genomes. That viral DNA always gets inserted in the same place, and new sequences get added after old ones, as if the bacteria were stacking books on a shelf. They use these archives to guide an enzyme called Cas9, which cuts and disables any viral DNA that matches the stored sequences.
So, first and foremost, CRISPR is a kind of genetic memory—a system for storing information. And that information doesn’t have to be the DNA of viruses. Scientists can now encode any digital file in the form of DNA, by converting the ones and zeroes of binary code into As, Cs, Gs, and Ts of the double helix.
By doing exactly that, Shipman and his colleagues encoded an image and a movie in a colony of bacteria. For the movie, he chose a simple five-frame GIF of Muybridge’s horse. “It was one of the first examples of a moving image, it was captured with a technology that was very new at the time, and it answered some relevant questions,” he says. And for the image, he chose a simple black-and-white photo of a hand. “It harkens back to the first images that humankind put in the natural world—handprints on cave walls,” he says. “We’re putting images into the natural world in a different way.”
Recording information in DNA isn’t new: That’s effectively what living things have been doing since the dawn of life itself. More recently, scientists have realized that DNA makes the perfect storage medium. It takes up so little space that you could fit all the world’s data in the back of a truck. It’s durable, provided it’s kept cold, dry, and dark. And it is immune to obsolescence: DVDs and Blu-Rays will eventually go the way of cassettes and laser discs, but humans will always have the desire and means to read DNA.
In 2011, George Church, Shipman’s supervisor, encoded his entire book in DNA, along with some images and a JavaScript program. A year later, two British researchers from the European Bioinformatics Institute used a more complex cipher to encode all of Shakespeare’s sonnets, a clip of Martin Luther King Jr.’s “I Have a Dream” speech, and a PDF of the paper from James Watson and Francis Crick that detailed the structure of DNA. More recently, Yaniv Erlich and Dina Zielinski, both from the New York Genome Center and Columbia University, used an even more efficient scheme to encode a silent movie, a computer operating system, a photo, a scientific paper, a computer virus, and an Amazon gift card.
All of these projects used naked DNA strands, floating within tubes of liquid. But other groups have written information into the genomes of actual living cells. In 1999, the artist Eduardo Kac encoded a sentence from the biblical book of Genesis into a microbe. In 2003, data scientist Pak Chung Wong stored the lyrics of the song “It’s a Small World” in the genomes of various bacteria, while another did the same for Einstein’s famous equation—E=mc²—in 2007. In 2010, Craig Venter and his team fashioned a bacterium with a fully lab-made genome, into which they had encoded their names and a quote from James Joyce: “To live, to err, to fall, to triumph, to re-create life out of life.” (This prompted an unhappy letter from Joyce’s estate.)
Shipman’s project began when he and his colleagues converted the image of a hand into DNA code, so that different sequences of DNA letters represented the color within each pixel. They manufactured all that DNA in the form of small strands, each of which was tagged to resemble the kind of viral sequences that the CRISPR system would naturally snag. The team delivered these strands to colonies of E. coli, which they grew overnight. They sequenced the part of the microbes' genomes where CRISPR information is stored, and decoded those sequences back into digital data. This allowed them to successfully recover the picture of the outstretched hand.
Encoding the horse GIF was more challenging. Not only did the team have to encode each frame, but also the order of the frames. Fortunately, CRISPR makes that easy. When bacteria grab viral DNA, they always insert new sequences after old ones, so the CRISPR system naturally orders information from newest to oldest. Shipman’s team took advantage of that. They offered their bacteria the DNA strands representing each frame of the GIF, one by one. By sequencing everything afterwards, they could recover the full movie. And if they loaded the bacteria with the DNA strands in the reverse order, they recovered a GIF of a horse running backwards. “We wanted to show that we didn’t need to encode timing information at all,” Shipman says.
Even when the team let the bacteria grow for a week—many generations in their time—the data didn’t degrade. “Those sequences aren’t doing anything, so there’s probably little cost to the having them,” says Shipman. Also, the data is distributed, so no single cell in the colony completely encodes the hand photo or the horse GIF. That prevents each individual bacterium from becoming overloaded with extraneous DNA.
“It’s a nice proof of principle of the potential for a living storage system,” says Dina Zielinski, who’s now at the Curie Institute in Paris. “Traditionally, the DNA in which data is encoded is finite. Once the data is written, it can be copied and read, like when you drag and drop new files onto your USB drive. But with a living system there's the potential to ‘write’ additional information. A living system allows for a more dynamic storage architecture.”
Your imagination could run wild thinking of applications for this. When I asked my colleagues, they came up with ideas like conducting cellular espionage by encoding secret messages in microbes, encoding your name in something else’s DNA like a form of genetic graffiti, and rickrolling future scientists by infusing bacteria with the lyrics to “Never Gonna Give You Up” and burying them in permafrost.
None of this is what Shipman has in mind. “We’re not using this to record movies,” he says. “We’re not going to put Wikipedia into bacteria.” Instead, he came into this as a neuroscientist who found it vexingly hard to study what happens as our brains develop—which genes are activated when, and where, and in which neurons? “It’s hard to reconstruct that because every time you touch the system, you disrupt it,” he says. “I imagined that if we had some way of encoding data in living cells while they’re still alive, and have it stored there, we could make progress.”
That’s the ultimate goal: turn cells into living recorders. “We want cells to go out and record environmental or biological information that we don’t already know.” That might include everything from the level of pollutants in a lake, or the genes that the microbes switch on as they go about their lives. All of this is a long way off, but the first necessary step is to show that cells can indeed store meaningful amounts of information.